MSPHATE
Abstracted biological features across all levels of data granularity
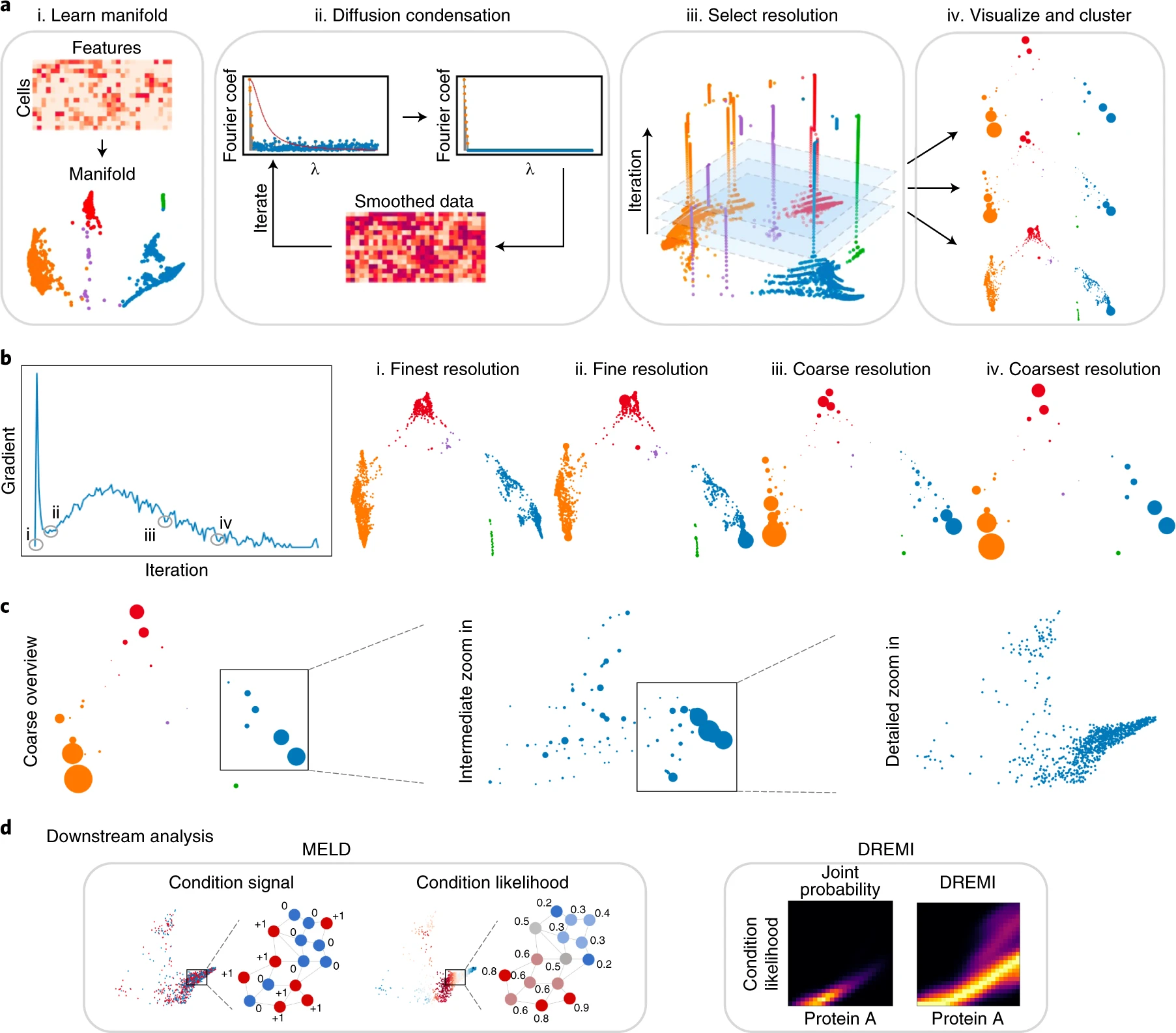
As the biomedical community produces datasets that are increasingly complex and high dimensional, there is a need for more sophisticated computational tools to extract biological insights. We present Multiscale PHATE, a method that sweeps through all levels of data granularity to learn abstracted biological features directly predictive of disease outcome. Built on a coarse-graining process called diffusion condensation, Multiscale PHATE learns a data topology that can be analyzed at coarse resolutions for high-level summarizations of data and at fine resolutions for detailed representations of subsets. We apply Multiscale PHATE to a coronavirus disease 2019 (COVID-19) dataset with 54 million cells from 168 hospitalized patients and find that patients who die show CD16hiCD66blo neutrophil and IFN-γ+ granzyme B+ Th17 cell responses. We also show that population groupings from Multiscale PHATE directly fed into a classifier predict disease outcome more accurately than naive featurizations of the data. Multiscale PHATE is broadly generalizable to different data types, including flow cytometry, single-cell RNA sequencing (scRNA-seq), single-cell sequencing assay for transposase-accessible chromatin (scATAC-seq), and clinical variables.